AI methods are developed and applied to medical imaging as part of infectious disease research at the IRF-Frederick. AI-guided preclinical models can be translated to human studies of disease.
In a collaborative effort with the Center for Infectious Disease Imaging (CIDI) at the NIH Clinical Center, post-doctoral and post-baccalaureate fellows develop novel machine-learning methods (such as, deep-learning-based medical image segmentation and disease state classification) toward the study of infectious diseases. The fellows are mentored by imaging scientists from the IRF-Frederick and radiologists from CIDI.
The AI Team works closely with the IRF-Frederick Imaging Sciences team and Chief Medical Officer to obtain multi-modality longitudinal medical imaging data and focus efforts on the most clinically relevant needs.
Main Areas of Focus
- Application of machine-learning methods for predictive analyses of infectious disease state and correlation with non-imaging biomarkers
- Segmentation of whole organs and abnormalities seen on multi-modal medical images
- Segmentation of histopathology slides using whole slide images
- Radiomic feature extraction from segmented images and application to machine-learning classification algorithms
- Use of image segmentation results from structural imaging applied to functional imaging (e.g., computed tomography [CT] to positron emission tomography [PET])
- Automated quantification of medical images for use as endpoints in infectious disease imaging research studies
- Quantitation of immunohistochemical-stained histopathology slides
- Correlation of histopathology slides with other markers of disease.
Capabilities
- Automated pipeline for deep-learning-based image segmentation
- Integration with high-performance computing environment (Locus), containing graphics processing unit (GPU) nodes for training and parallel execution of AI methods
- Multi-organ segmentation (such as, liver, spleen, lungs, lung lobes, and lung lesions)
- Creation of ground-truth annotations
- Continuous improvements to deep-learning-based segmentation models
- Collaboration with other IRF-Frederick teams (including Pathology and Histology, Immunology, and data management) to integrate multi-parameter estimations into machine-learning methodologies
- Generalize specific segmentation models for use by external collaborators through the development of software tools for image data augmentation
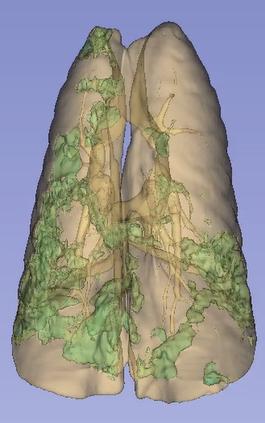
Three-dimensional rendering of deep-learning-based segmentation of lungs and lung infiltrates (green), caused by viral infection.

Automated deep-learning-based segmentation of liver (green outline) and spleen (pink outline) in computed tomography (CT) on the left and positron emission tomography (PET) on the right of a nonhuman primate.
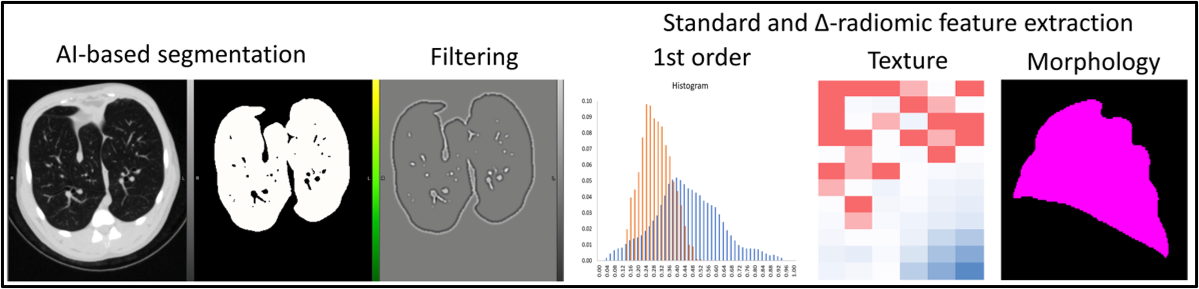
Radiomic feature extraction (1st order, texture, and morphology) from segmented lungs on computed tomography (CT) image and correlation matrix of features.
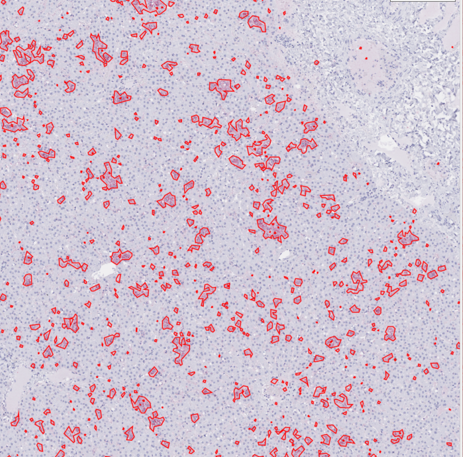
Segmentation for quantification of immunohistochemical stain for viral glycoprotein in liver slide from nonhuman primate. Viral glycoprotein is visible after staining and subsequently outlined via segmentation. Each outlined glycoprotein deposition may be an area of interest for further examination.
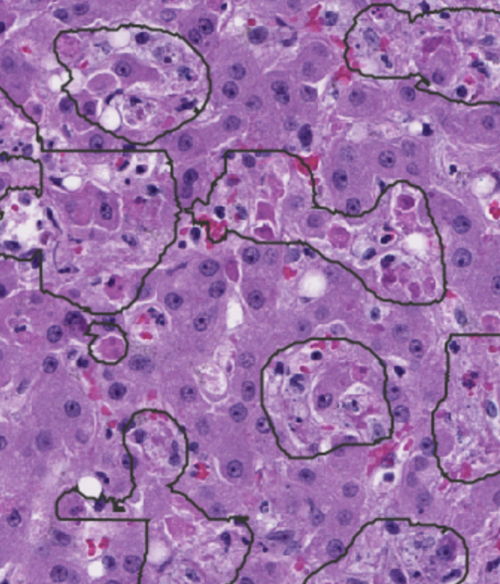
Automated machine-learning-based segmentation in liver slides. A trained segmentation script allows for the defining of regions of interest (outlined) that include dead/dying cells caused from viral infection.
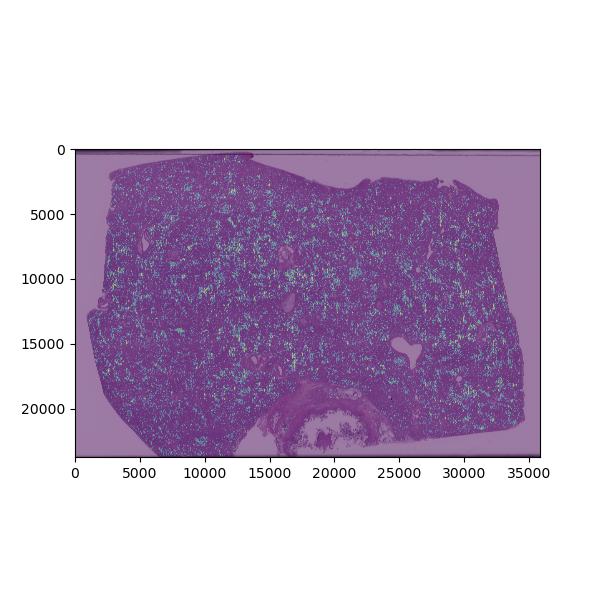
Clustering analysis based on morphologic features of necrotic areas in liver slide from viral infection. After machine-learning-based algorithms are used to identify areas of necrosis, visual features of these areas are extracted and used to differentiate types of necrosis into clusters. The plot is color-coded to depict how each necrotic area is grouped based on this clustering analysis.
Select Publications
Through collaborative work, the AI Team has played a critical role in a number of publications. Peruse a list of papers and conference posters.
Location
Integrated Research Facility at Fort Detrick (IRF-Frederick)
Contact Information
Jeffrey Solomon, Ph.D.
Artificial Intelligence Lead
Senior Imaging Scientist (Contractor)
C. Paul Morris, M.D., Ph.D.
Senior Pathology and Informatics Scientist
IRF-Frederick
Standards
All procedures are well-documented and adhere to standard operating procedures (SOPs), methods, or study-approved plans and agreements.
Collaboration Opportunities
- Studies relevant to human disease
- Use of surrogate systems to test clinical hypotheses
- Use of biological systems to answer questions regarding disease pathogenesis and strategies for intervention including antimicrobials, vaccines, and other countermeasures
- Developing and incorporating cutting-edge technologies to understand infectious diseases
Read more about how to work with the IRF-Frederick.

