An Analysis: Evolutionary Distance of SARS-CoV-2 and Bat Coronaviruses Studied Under the NIH-supported Research Grant to EcoHealth Alliance
The research that NIH approved under the grant to EcoHealth Alliance with a subaward to the Wuhan Institute of Virology in Wuhan, China sought to understand how animal coronaviruses, especially bat coronaviruses, evolve naturally in the environment and have the potential to become transmissible to the human population. This research included studying viral diversity in bat reservoirs, surveying people who work in live animal markets or other occupations with high exposure to wildlife for evidence of bat coronavirus infection and analyzing data to predict which newly discovered viruses pose the greatest threat to human health.
Coronaviruses use a protein called spike to bind to a protein on the surface of a host cell to facilitate infection. Some coronaviruses, including SARS-CoV-1 (the cause of the SARS outbreak in 2003) and SARS-CoV-2 (the cause of the COVID-19 pandemic), use the angiotensin converting enzyme-2 (ACE2) protein to help enter and infect host cells. In order to study animal coronaviruses circulating in nature, the investigators replaced the spike protein from a well-characterized bat coronavirus, WIV1-CoV, with the spike protein of animal coronaviruses recently discovered in bats in China. Using techniques common in virology, experiments involved a single round of infection in several cell lines, and in some cases, in mice that were genetically modified to express the human version of ACE2. All other aspects of the mice, including the immune system, remained unchanged. The ACE2 transgenic mice were used to determine if spike proteins from bat coronaviruses discovered in China were capable of binding human ACE2, and therefore, whether the bat coronaviruses themselves, which were already present in the environment, could potentially infect humans and cause disease. WIV1-CoV is not known to cause infection in humans but has been shown in the laboratory to infect both human cells and ACE2 transgenic mice (ref), making it an ideal tool to use for these studies. Several of the bat coronaviruses used in these experiments were also found to be capable of replicating in ACE2 transgenic mice, indicating that the spike protein from the naturally occurring bat coronaviruses from which they were made could bind ACE2 in vivo.
Questions have been raised about whether this NIH-funded research had a role in the emergence of SARS-CoV-2. In this regard, the chimeric viruses that were studied (i.e., the WIV-1 virus with the various spike proteins obtained from bat viruses found in nature) were so far distant from an evolutionary standpoint from SARS-CoV-2 (Figure 1) that they could not have possibly been the source of SARS-CoV-2 or the COVID-19 pandemic. The body of the scientific data from this award including the bat coronavirus sequences published in the scientific literature and public databases makes this conclusion readily apparent to anyone with experience in and knowledge of virus phylogeny and evolutionary biology.
Figure 1. Relationship of bat coronaviruses to SARS-CoV-1 and SARS-CoV-2.
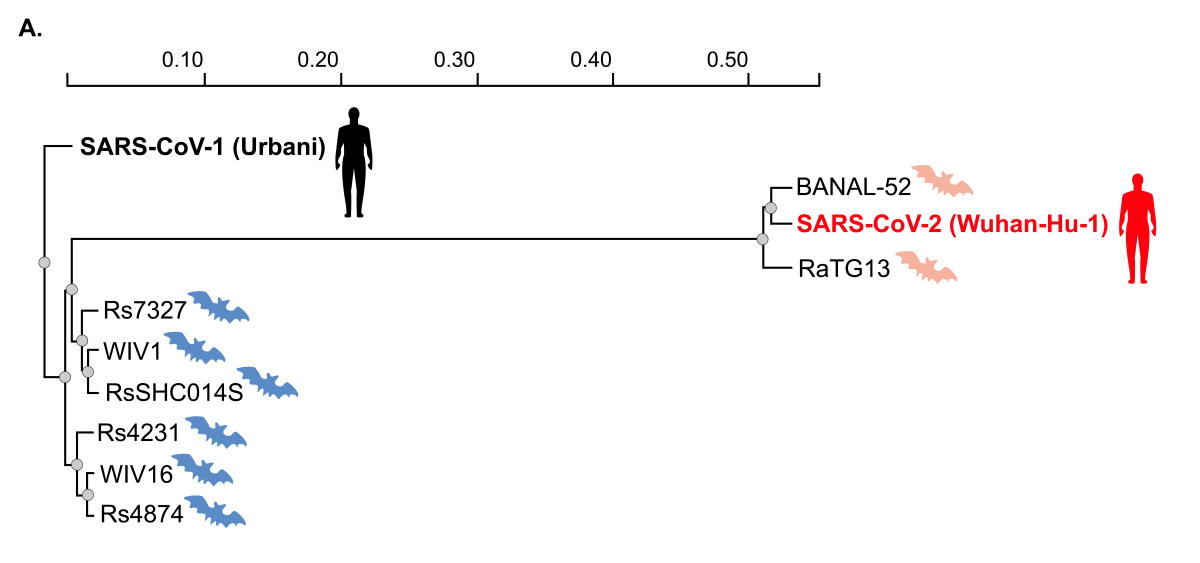
A) A phylogenetic tree based on nucleotide sequences of indicated coronavirus spike proteins demonstrating the evolutionary distance of SARS-CoV-2 with the bat coronaviruses experimentally studied under the NIH grant to EcoHealth Alliance (blue bat icons). Bat coronaviruses most closely related to SARS-CoV-2, none of which were studied in the EcoHealth grant, are denoted with orange bat icons. The scale bar represents the number of nucleotide substitutions per site.
B) Comparison of the nucleotide sequence identity of indicated coronaviruses to SARS-CoV-2. The left panel shows the percent identity of indicated coronavirus spike nucleotide sequences to SARS-CoV-2. The right panel shows the percent nucleotide identity of the indicated full coronavirus genomes to SARS-CoV-2.
Despite the similarity of RaTG13 and BANAL-52 bat coronaviruses (orange bars) to SARS-CoV-2 (red bars), experts agree that even these viruses are far too divergent to have been the progenitor of SARS-CoV-2, further highlighting that the bat coronaviruses studied under the EcoHealth Alliance grant (blue bars) could not have been the source of SARS-CoV-2 and the COVID-19 pandemic. Several other similarly divergent viruses that failed to replicate in cells are not shown (ref).
The above figure shows the sequence relationships between SARS-CoV-1, SARS-CoV-2 and the naturally occurring bat coronaviruses used in experiments under the NIH grant to EcoHealth Alliance and reported in the scientific literature (ref) or annual progress reports. From this analysis, it is evident that the viruses studied under the EcoHealth Alliance grant are very far distant from SARS-CoV-2. Included for comparison is RaTG13, one of the closest bat coronavirus relatives to SARS-CoV-2 collected by the Wuhan Institute of Virology (ref) and BANAL-52, one of several bat coronaviruses recently identified from bats living in caves in Laos (ref). Although RaTG13 and BANAL-52 are 96-97% identical to SARS-CoV-2 at the nucleotide level (>900 nucleotide differences across the entire genome), the difference actually represents decades of evolutionary divergence from SARS-CoV-2. Experts in evolutionary biology and virology have made it clear that even the closest known relatives of SARS-CoV-2, which were not studied under the EcoHealth Alliance grant, are evolutionarily too distant from SARS-CoV-2 to have been the progenitor of the COVID-19 pandemic (ref, ref). Field studies continue the search for more proximate progenitors.


