Viral Biochemistry Section
Established in 2000
Klaus Strebel, Ph.D.
Chief, Viral Biochemistry Section

Major Areas of Research
- Biological and biochemical functions of HIV and SIV accessory proteins Vif, Vpu, Vpr, and Vpx
- Characterization of virus-host interactions
- Characterization of cellular factors controlled by Vif, Vpu, Vpr, and Vpx
- Characterization of innate immune defense mechanisms
Program Description
Primate immunodeficiency viruses, including HIV-1, are characterized by the presence of accessory genes such as vif, vpr, vpx, vpu, and nef. Of note, none of these proteins have enzymatic activity. Instead, they act as molecular adaptors to either redirect the normal function of the host factors for virus-specific purposes or to inhibit their normal physiological function by targeting proteins for degradation or by altering their intracellular localization.
The primary objective of the Viral Biochemistry Section (VBCS) is to gain insights into innate host defense mechanisms and to decipher the molecular mechanisms governing the functional neutralization of innate restriction factors by HIV- or SIV-encoded proteins. The ultimate goal is to identify possible points of interference that could lead to the development of novel treatment strategies. Over the last few years, our work on Vif has focused on characterizing mechanisms of dominant-negative interference by defective Vif variants. We also performed a comparative characterization of Vif proteins from HIV-1 versus HIV-2 and observed an interesting difference in how the two viruses deal with APOBEC3B (A3B) restriction.
In a separate line of research, we studied the effects of HIV-1 infection of primary macrophage cultures. We found that HIV-1 virus production from infected macrophages is inhibited by mannose receptor 1 (MRC1). Interestingly, we found evidence to suggest that MRC1 not only affects the release of mature virions from infected macrophages but, in addition, it can also affect the infectivity of the viruses produced in the presence of MRC1. Of note, we found that HIV-1 was able to counteract this effect through transcriptional silencing of the MRC1 promoter in a Tat-dependent manner. We are currently defining the precise molecular mechanisms involved in the control of MRC1 expression in primary macrophages. In particular, we are interested in characterizing a potential role of HIV-1 Vpr in this process.
In a third line of research, we characterized a novel, myeloid cell-specific Vpr-interacting protein, gelsolin. We found that gelsolin reduced expression and packaging of HIV-1 Env into viral particles, thereby reducing viral infectivity. This effect was inhibited by the HIV-1 Vpr protein, which caused degradation of gelsolin. Interestingly, pseudotyping studies revealed that gelsolin was also able to inhibit the expression and packaging of the VSVg glycoprotein into HIV-1 particles. These results suggest that gelsolin may promote lysosomal turnover not only of HIV-1 Env but of other cell-surface glycoproteins as well.
Aside from that. we are currently characterizing two additional restriction factors to understand their antiviral action at the molecular level. Finally, we are continuing our international collaborative effort aimed at gaining a better understanding of the HIV-2 epidemiology, drug resistance, and pathogenesis in Cape Verde, an archipelago located offshore from Senegal at the West African coast with an unusually high incidence of HIV-2 infections. This will involve the functional characterization of HIV-2 accessory genes isolated form primary HIV-2 isolates in primary human cells.
Biography
Education
Ph.D., 1985, University of Heidelberg, GermanyDr. Strebel received his Ph.D. in microbiology in 1985 from the University of Heidelberg, Germany. After postdoctoral research in Germany on foot-and-mouth disease protein processing and maturation, he joined the Laboratory of Molecular Microbiology (LMM) in 1986 as a postdoctoral fellow to work on molecular mechanisms of HIV-1 replication. He was awarded tenure in 1998 and, since 2000, has been chief of the Viral Biochemistry Section within LMM.
Selected Publications
Helena Fabryova, Sayaka Sukegawa, Eri Miyagi, Louis Taylor, Sandra Kao, Damien Ferhadian, Heiner Schaal, Frank Hillebrand, Klaus Strebel. HIV-1 Vpr induces degradation of Gelsolin, a myeloid cell-specific host factor that reduces viral infectivity by inhibiting expression and packaging of the HIV-1 Env glycoprotein. mBio (2022 revised)
Kao S, Miyagi E, Mallorson R, Saito H, Sukegawa S, Mukherji A, Mateja A, Ferhadian D, Fabryova H, Clouse K, Strebel K. The Myeloid-Specific Transcription Factor PU.1 Upregulates Mannose Receptor Expression but Represses Basal Activity of the HIV-LTR Promoter. J Virol. 2022 Jun 29:e0065222.
Bandarra S, Miyagi E, Ribeiro AC, Gonçalves J, Strebel K, Barahona I. APOBEC3B Potently Restricts HIV-2 but Not HIV-1 in a Vif-Dependent Manner. J Virol. 2021 Nov 9;95(23):e0117021.
Miyagi E, Welbourn S, Sukegawa S, Fabryova H, Kao S, Strebel K. Inhibition of Vif-Mediated Degradation of APOBEC3G through Competitive Binding of Core-Binding Factor Beta. J Virol. 2020 Mar 17;94(7):e01708-19.
Fabryova H, Strebel K. Vpr and Its Cellular Interaction Partners: R We There Yet? Cells. 2019 Oct 24;8(11):1310.
Sukegawa S, Miyagi E, Bouamr F, Farkašová H, Strebel K. Mannose Receptor 1 Restricts HIV Particle Release from Infected Macrophages. Cell Rep. 2018 Jan 16;22(3):786-795.
Recent Progress
Mannose receptor inhibits virus release from HIV-infected macrophages
Human mannose receptor 1 (MRC1) is expressed on the surface of most tissue macrophages, dendritic cells, and select lymphatic or liver endothelial cells. MRC1was reported to serve as entry receptor for invading pathogens such as bacteria, fungi, viruses (incl. HIV-1), and other parasites. Interestingly, the MRC1-mediated uptake of HIV-1 by macrophages does not lead to productive infection but results in the processing in MHC-II-containing compartments for subsequent antigen presentation. Thus, MRC1 constitutes part of a cellular innate host defense mechanism.
It was reported that productive HIV-1 infection of macrophages results in the downmodulation of MRC1. However, nothing was known about the effect of MRC1 on virus replication. Our overall interest in the HIV-induced modulation of cell surface markers such as CD4 or BST-2 and their functional consequences for virus replication prompted us to examine the functional correlation between MRC1 expression on primary human macrophages (MDM) and productive HIV-1 replication.
We found that silencing of MRC1 in MDM resulted in enhanced virus production similar to what was previously observed in response to the HIV-1 Vpu-induced antagonism of BST-2. Conversely, overexpression of MRC1 following transient transfection of HEK293T cells resulted in significant inhibition of particle release (Figure 1). Virions produced in the presence of MRC1 accumulated in clusters at the cell surface. These virions, when detached from the cells, were fully infectious. Despite its phenotypical similarity to BST-2-mediated restriction of virus release, MRC1-dependent inhibition of virus production was BST-2 independent because: (1) it was observed in HEK293T cells, which are inherently BST-2-negative; and (2) the presence of Vpu had no impact on MRC1-dependent inhibition of virus release. Surprisingly, deletion of Env did not alleviate the MRC1-imposed inhibition of virus release, suggesting the involvement of additional cellular factor(s) in the process (Figure 2). Overall, our data suggest that MRC1 represents an antiviral factor affecting virus release in a BST-2-like but mechanistically distinct manner.
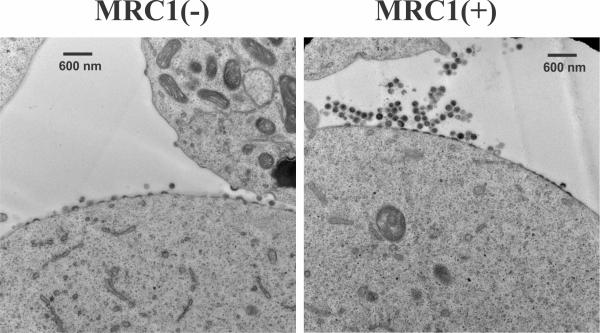
Fig. 1: Expression of hMRC1 results in the accumulation of clusters of viral particles at the cell surface. HEK293T cells were transfected with pAD8 together with empty vector (left) or pCMV6-hMRC1 (right) and cultured for 32 hr. Cells were then processed for transmission EM.
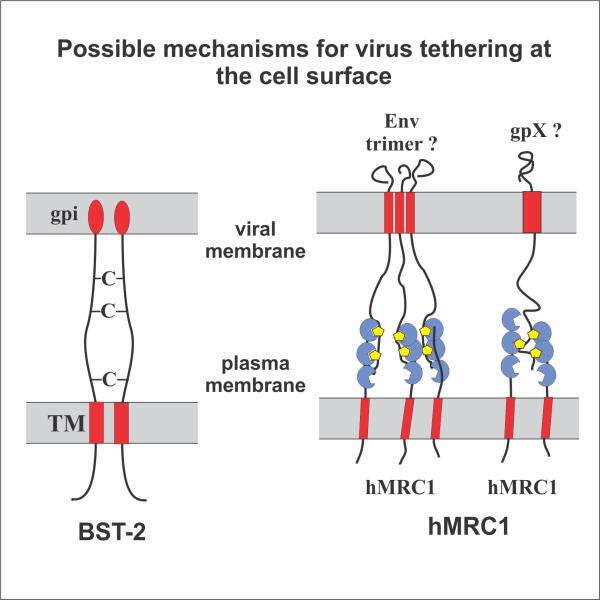
Fig. 2: Retention of virus particles by BST-2 (left) and hMRC1 (right). BST-2 tethers virions via its two membrane anchors (red). hMRC1 retains viral particles via its carbohydrate recognition domains (blue) that attach to carbohydrate moieties (yellow) on virus-associated glycoproteins (e. g. HIV-1 Env).
Publications arising from this work
Sukegawa S., Miyagi E., Bouamr F., Farkasova H., and Strebel K. Mannose Receptor 1 restricts HIV virus release from infected macrophages. Cell Reports 22(3):786-795 (2018).
HIV-1 Tat induces transcriptional silencing of the mrc1 promoter.
Some pathogens, including HIV-1, have evolved to antagonize the innate immune function of MRC1 by downregulating mannose receptor from the cell surface. In the case of HIV-1, the mechanism of MRC1 downmodulation is still under debate. Nef, Vpr, and Tat were all implicated in this effect. Nef was reported to induce cell surface downmodulation of MRC1 without affecting its steady-state levels while Vpr was reported to induce degradation of MRC1. Finally, HIV-1 Tat was reported to inhibit transcription from the rat MRC1 promoter, a finding that was not confirmed by others. Thus, the precise mechanism of HIV-induced down-modulation of MRC1 from the surface of productively infected macrophages remained unclear.
Our own experiments suggest inhibition of MRC1 at the transcriptional level. Indeed, individual deletion of any of the viral accessory proteins did not interfere with the HIV-induced downmodulation of MRC1. To directly test the impact of Tat on MRC1 expression, we cloned a 150 bp fragment of the human MRC1 promoter into the promoter trap vector pGL3 (pGL-hMRC1-Luc; Figure 3A). Unexpectedly, transfection of pGL-hMRC1-Luc together with Tat neither activated nor inhibited luciferase expression (not shown). Inspection of the MRC1 promoter sequence revealed the presence of several transcription factor binding sites, including two copies of a putative binding site for the myeloid-specific transcription factor PU.1. Indeed, when co-transfecting PU.1 vector with pGL-hMRC1-Luc, we observed a dose-dependent increase in luciferase activity (Figure 3B). Of note, when increasing amounts of Tat were transfected together with constant amounts of PU.1, we observed a dose-dependent inhibition of luciferase activity (Figure 3C). Thus, HIV-1 Tat acts as a PU.1-dependent transcriptional inhibitor of the MRC1 promoter. The same result was obtained when full-length proviral vectors were tested. The inhibitory effect of Tat on MRC1 promoter activity is contrasted by its well-known activating effect on the HIV-1 LTR promoter. The HIV-1 LTR does not have any classical PU.1 binding sites.
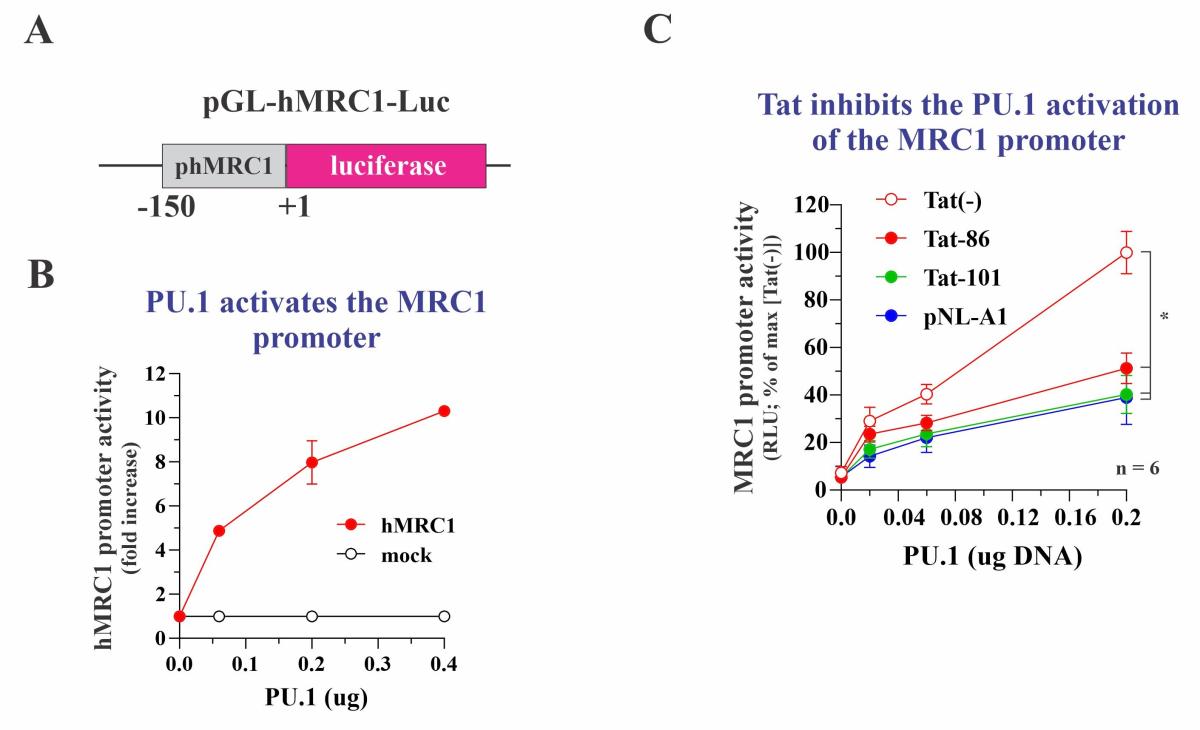
Fig. 3: Tat inhibits the PU.1 dependent activation of the MRC1 promoter. (A) Schematic of the test vector. (B) PU.1 activates the MRC1 promoter in a dose-dependent manner. (C) Expression of various forms of Tat inhibit the PU.1 activated MRC1 promoter. Tat-86 and Tat-101 are two forms of AD8 Tat. pNL-A1 expresses NL4-3 Tat.
We nevertheless tested the effect of PU.1 on the transcriptional activity of the HIV-1 LTR promoter. For that purpose, a roughly 600 bp fragment from the NL4-3 5’-LTR was cloned into the pGL3-Luc vector (Figure 4A). As expected, the LTR-Luc vector had significant basal activity, which was increased by more than 10-fold in the presence of Tat. Intriguingly, co-expression of PU.1 induced a dose-dependent inhibition of the LTR-mediated luciferase expression both in the absence and presence of Tat (Figure 4B). Thus, PU.1 behaved as a transcriptional activator of the MRC1 promoter but acted as a transcriptional inhibitor of the HIV-1 LTR promoter in a Tat independent manner. Importantly, siRNA-mediated silencing of PU.1 in HIV-infected MDM resulted in increased HIV-1 gene expression, indicating that the inhibitory effect of PU.1 on HIV-1 transcription can be observed in HIV-infected primary human macrophages and is therefore functionally relevant. We propose a model of a regulatory circuit, in which PU.1 inhibits the HIV-1 LTR resulting in reduced expression of Tat (Fig. 5:c). However, Tat can increase its own expression by activating the HIV LTR (Fig. 5:d). Increased Tat levels result in reduced levels of PU.1 (Fig. 5:e), which antagonizes its effect on the HIV LTR (Fig. 5:c) and at the same time reduces MRC1 promoter activity (Fig. 5:b). Reduced MRC1 levels then lead to increased viral particle release (Fig. 5:a). Thus, HIV-1 Tat, through its effect on PU.1 increases HIV-1 gene expression and at the same time facilitates the release of viral particles from infected macrophages via inhibition of MRC1 expression.
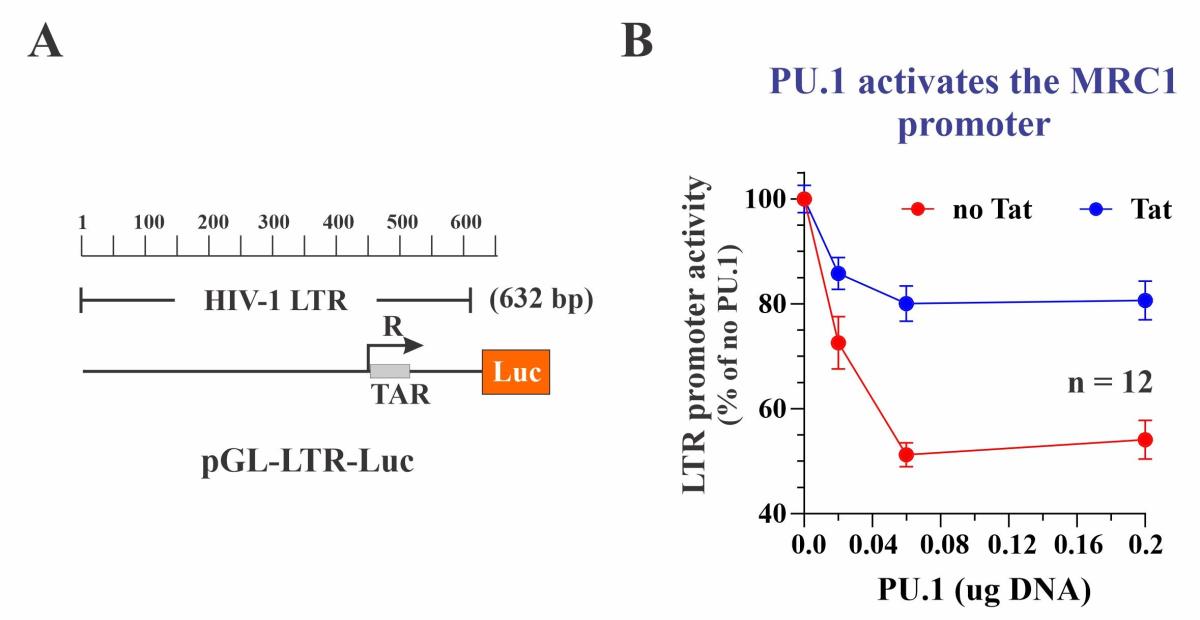
Fig. 4: PU.1 inhibits the HIV-1 LTR. (A) Diagram of the indicator plasmid used. (B) PU.1 inhibits LTR promoter activity +/- Tat in a dose-dependent manner.
Publications arising from this work
Kao et al 2022 (in press).
Proposed Course of Work
We will continue our work on PU.1 and MRC1 expression. We have mutated or deleted the two putative PU.1 binding sites on the MRC1 promoter element either alone or in combination. Preliminary data indicate that the integrity of one of the two binding sites is sufficient for PU.1 responsiveness. We will perform electrophoretic mobility shift assays (EMSA) to determine binding of PU.1 and Tat to the various promoter elements. In particular, we will investigate if PU.1 binding to the HIV-1 LTR is required and if so, try to identify the binding site. We will also transfer PU.1 binding sites into heterologous, PU.1 unresponsive, promoters to determine the minimal sequences required for PU.1 responsiveness. PU.1 has an N-terminal transcriptional activation domain, a central PEST sequence, and a C-terminal DNA binding domain. We will study PU.1 mutants to determine whether the same domains are required for transcriptional activation and inhibition.
![Figure 5: PU.1 inhibits transcription from the HIV-1 LTR but activates the MRC1 promoter. PU.1 is a myeloid-specific transcription factor that regulates expression of multiple genes, including mannose receptor (MRC1). PU.1 mediated upregulation (blue arrow) of MRC1 [a] (see Fig. 3B) accounts for the previously observed inhibition of HIV-1 virus release [b]. This effect is counteracted by the HIV-encoded Tat protein that inhibits PU.1 function [c] (see Fig. 3C). Inhibition of PU.1 function by Tat reduces MRC](/sites/default/files/styles/image_style_100_width_xs/public/2022-update-fig-5.jpg?itok=5hfaoKN3)
Figure 5: PU.1 inhibits transcription from the HIV-1 LTR but activates the MRC1 promoter. PU.1 is a myeloid-specific transcription factor that regulates expression of multiple genes, including mannose receptor (MRC1). PU.1 mediated upregulation (blue arrow) of MRC1 [a] (see Fig. 3B) accounts for the previously observed inhibition of HIV-1 virus release [b]. This effect is counteracted by the HIV-encoded Tat protein that inhibits PU.1 function [c] (see Fig. 3C). Inhibition of PU.1 function by Tat reduces MRC1 expression, which contributes to enhanced virus release. While PU.1 upregulates MRC1 expression, it has an inhibitory effect on HIV-1 LTR promoter activity [d] (see Fig.4B). This leads to reduced Tat protein synthesis [e], which in turn increases the inhibition of HIV-1 LTR transcription by PU.1 [d]. However, a positive feedback loop ensures continued Tat synthesis via TAR-dependent activation of HIV-1 transcription [f] and thus control of PU.1 expression. Aside from Tat, HIV-1 Vpr and Nef have been implicated in the regulation of MRC1 [g].
Research Group
Molecular and virological investigation of HIV-1 and HIV-2 and their interactions with host factors.


