Computational Systems Biology Section
Thorsten Prüstel, Ph.D.
Staff Scientist

Major Areas of Research
- Stochastic simulation approaches to cellular signaling
- Computational models of cellular morphology
- Interplay between stochastic and spatial aspects of cellular signaling at cell-cell contacts (example: T-cell receptor activation)
- Interactions between migrating cells and their environment
Program Description
A major focus of our research is on developing computational modeling approaches that are not only able to predict the behavior of complex biological systems, such as the human immune system, but that also provide insights into the mechanisms that underlie the systems’ behavior. Such a mechanistic understanding of those cellular processes that play key roles in health and disease is critical in the search of novel treatment strategies. It requires simulation methods that can describe the intricate interactions of the systems’ components on a variety of biological scales, ranging from single molecules, over individual cells and multicellular structures to whole organisms.
On the most fundamental biological scale that our research covers, the level of single molecules and reaction-diffusion driven interactions of molecular complexes, subcellular processes behave in a stochastic fashion. Therefore, an important research objective has been the development of high-resolution single-particle stochastic simulation methods that can correctly and efficiently capture reaction-diffusion processes in the context of cellular biochemistry, for instance at and adjacent to cell surfaces, where many of the cellular signaling processes are initiated. Accordingly, a further major research goal has been the development of computational representations of arbitrarily shaped geometries. Here, a focus has been on models that are flexible enough to not only represent the shape of separate cells, but also interfaces between two cells. Such interfaces form, for instance, when two immune system cells exchange information on pathogens (see figure below).
The availability of efficient and detailed stochastic simulations is a prerequisite for establishing a bridge between the stochastic events of cellular biochemistry on the molecular scale and the higher-level cellular behavior that underlies health and disease.
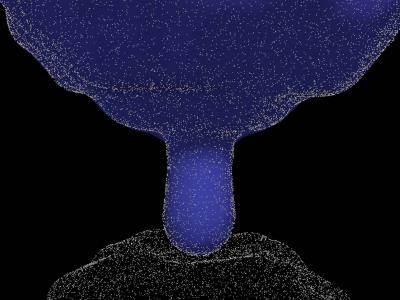
Snapshot of a high-resolution single-particle stochastic simulation of a T-cell (blue-colored model) and an antigen-presenting cell interacting with each other via a T-cell protrusion contact. Bright dots represent single T-cell receptors and peptide: MHC complexes.
Biography
Education
B.S., Ph.D., University of Hamburg, Germany
Languages Spoken
GermanThorsten Prüstel received his B.S. and Ph.D. in theoretical physics from the University of Hamburg in Germany. After conducting post-doctoral research under the supervision of Dr. Martin Meier-Schellersheim, he became a staff scientist in the Computational Systems Biology Section.
The main foci of Thorsten’s research are on stochastic modeling and quantitative analyses of interactions between cells and their environment. The goal of his research is to contribute to a better understanding of how molecular mechanisms allow the immune system to process and categorize information about pathogens it encounters. Realizing that all molecular interactions underlying cellular signaling processes are inherently stochastic, Thorsten has been developing mathematical approaches and computational tools for single particle-based modeling of biological reaction-diffusion processes.
A major contribution of his research is the first derivation of the two-dimensional Green’s function describing stochastic interactions on membranes. Thorsten is the main developer of the stochastic component of the modeling software Simmune for which he is integrating single particle-based representations of cellular signaling pathways with realistic models of cellular morphology.
Selected Publications
Prüstel T, Meier-Schellersheim M. Space-time histories approach to fast stochastic simulation of bimolecular reactions. J Chem Phys. 2021 Apr 28;154(16):164111.
Johnson ME, Chen A, Faeder JR, Henning P, Moraru II, Meier-Schellersheim M, Murphy RF, Prüstel T, Theriot JA, Uhrmacher AM. Quantifying the roles of space and stochasticity in computer simulations for cell biology and cellular biochemistry. Mol Biol Cell. 2021 Jan 15;32(2):186-210.
Prüstel T, Meier-Schellersheim M. Unified path integral approach to theories of diffusion-influenced reactions. Phys Rev E. 2017 Aug;96(2-1):022151.
Prüstel T, Tachiya M. Reversible diffusion-influenced reactions of an isolated pair on some two dimensional surfaces. J Chem Phys. 2013 Nov 21;139(19):194103.
Prüstel T, Meier-Schellersheim M. Exact Green's function of the reversible diffusion-influenced reaction for an isolated pair in two dimensions. J Chem Phys. 2012 Aug 7;137(5):054104.
Angermann BR, Klauschen F, Garcia AD, Prüstel T, Zhang F, Germain RN, Meier-Schellersheim M. Computational modeling of cellular signaling processes embedded into dynamic spatial contexts. Nat Methods. 2012 Jan 29;9(3):283-9.

