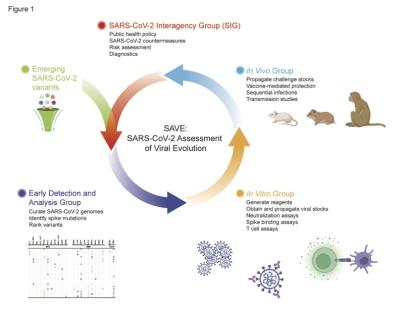
When the next SARS-CoV-2 variant of concern emerges in the world, NIAID will call on SAVE—its version of The Avengers—to quickly react and protect people. SAVE—which stands for SARS-CoV-2 Assessment of Viral Evolution—members assess whether mutations in variants can affect virus transmission, severity and immunity; test vaccines and therapeutics; and guide responses.
Like the comic book heroes fighting various mutant threats, SAVE focuses on mutations in SARS-CoV-2 and emerging virus variants. But program members say the global collaborative concept is a broad model for rapidly responding to evolving pathogens with pandemic potential.
NIAID assembled and coordinated SAVE scientists in January 2021, drawing on experts from around the world who specialize in relevant research fields such as viruses, the immune system, vaccines, epidemiology, structural biology, bioinformatics, virus genetics, and evolution.
SAVE members represent 58 different research sites located in the United States and around the world. Members participate within three sub-groups based on their expertise:
- Early Detection and Analysis
- In Vitro – what they can learn using flasks, beakers and tubes
- In Vivo – what they can learn in animal models that mimic human disease
“Collaboration within and across these groups has accelerated research and discovery due to immediate and open sharing of ideas, reagents, protocols and data,” 129 SAVE members said in a program overview just published in Nature.
SAVE emerged from the U.S. Department of Health and Human Services’ SARS-CoV-2 Interagency Group (SIG). The repeated emergence of COVID-19-causing variants of concern made SIG leaders recognize a critical need for scientists to rapidly generate, share and assess variant data in a highly coordinated manner to enable decision-making for public health.
The most telling example is how SAVE responded when the Omicron variant emerged in fall 2021. The group rapidly generated virus plasmids and spike protein; isolated, propagated and distributed authentic Omicron virus for research; submitted reagents to public repositories; performed binding and neutralization tests; and evaluated virus infection in different animal models.
Researchers then shared data from these studies with government agencies and submitted them as manuscripts online prior to peer-review—something rarely done before the COVID-19 pandemic. “The head-to-head comparison, review and discussion of unpublished data has yielded real-time peer review that would otherwise take months to achieve,” their summary states.
Each of the three groups has had to face various challenges in prioritizing work. For example:
- How much data is needed to provide relevant results in a desired, rapid timeframe? How can analysis be adjusted to account for low levels of sequencing in some countries and high levels in others?
- How can immune responses be enhanced to contribute to protection from infection and severe disease, and increase durability of immunity?
- Which animal models are best to evaluate protective immunity against a variant? Does immunity to variants differ in those who were infected after being vaccinated and those reinfected after having infection-induced immunity?
Regardless of challenges, the SAVE group sees a great need and opportunity for its type of collaboration in responding to outbreaks beyond the current pandemic.
“Over the past two decades, we have witnessed the emergence\re-emergence of several RNA viruses, including West Nile virus, H1N1 influenza virus, chikungunya virus, Zika virus, SARS-CoV-1, MERS-CoV, and Ebola virus, that have threatened global public health,” their summary states. “Developing collaborative programs between academic, industry and commercial partners is essential to respond to rapidly evolving viruses.”
Reference: M DeGrace, et al. Defining the risk of SARS-CoV-2 variants on immune protection. Nature DOI: 10.1038/s41586-022-04690-5 (2022).