In the early 1980s, scientists identified clumps of abnormal, misfolded prion protein in mammals as the cause of brain-wasting diseases, now called prion diseases. Since that time, they have struggled to answer: What does a normal prion protein do?
The answer, they believe, could help lead them to develop treatments and disease-prevention measures against human prion diseases, such as Creutzfeldt-Jakob disease, fatal familial insomnia and kuru, as well as animal prion diseases, such as scrapie in sheep and chronic wasting disease in cervids.
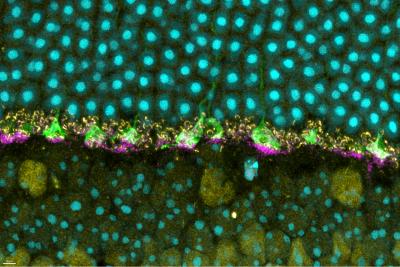
Now, a new study published in iScience from NIAID scientists at Rocky Mountain Laboratories in Hamilton, Montana, and colleagues provides details of how prion protein functions in the retina of mouse eyes, helping them respond to light.
The scientists used mice specially bred without prion protein to compare to wild mice with natural prion protein. Prior studies have suggested that prion protein may have a role in how nerves transmit signals to other nerves at specialized junctions, known as neural synapses. So, knowing that prion protein exists naturally in the eye, the researchers examined mouse retina for a specific neural synapse role.
A key tool the researchers used involved measuring the electroretinographic (ERG) responses – the amount of time it took for the retina in mice to respond to a flash of light. Remember as a kid in school learning about rods and cones in the eye and how they convert light signals to help the brain understand vision? The same is true in mice.
Compared to the wild mice with prion protein, the scientists observed deficiencies in ERG responses for mice without prion protein. The deficiencies affected the normal function of the rods and cones. And – using the ERG data and neural synapse information – they found that the deficiencies originated in the portion of the retina where natural prion protein was most highly concentrated.
Though additional study is needed, the researchers believe the prion protein may act like scaffolding to help cells and elements of the eye, such as rods and cones, to stabilize neural synapses. And they believe prion protein must be present for rods and cones to function normally.
The research team hopes these findings help colleagues who study prion diseases better understand what might occur in humans when natural forms of prion protein are therapeutically removed. New treatment strategies for prion diseases focus on using drugs that remove natural prion protein to eliminate the potential for misfolding and clumping. But researchers do not know whether that could result in unwanted outcomes, such as possibly affecting vision. These findings also could extend to other protein-related neurodegenerative diseases, such as Alzheimer’s (amyloid beta protein) and Parkinson’s diseases (alpha synuclein protein).
Scientists from Duke University and the McLaughlin Research Institute in Great Falls, Montana, collaborated on the study.
Reference: J Striebel, et al. The prion protein is required for normal responses to light stimuli by photoreceptors and bipolar cells. iScience DOI: 10.1016/j.isci.2024.110954 (2024).