NIAID-led Work Identifies Bacteria Signaling for Nerve Repair in Mice
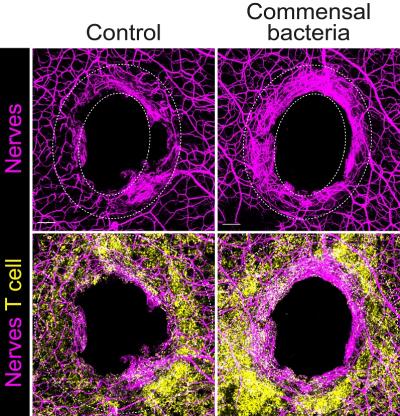
A team of NIAID-led researchers has identified a mechanism in mice in which the immune system and commensal bacteria – microbes that naturally colonize tissues – help repair damaged sensory neurons within the skin. They hope their findings, published in Cell, could lead to therapies that stimulate recovery in people following skin injury and limit damage from chemotherapy and chronic diseases, such as diabetes.
When commensal bacteria colonize the skin, they fine-tune the immune response – known as adaptive immunity – without representing any threat. This study found that when an injury occurs to a colonized surface, say a skin puncture, the preemptive immunity established from the commensal bacteria can help the host recover damaged sensory neurons. That means recovering awareness to touch, temperature, pain and itch.
The mechanism involves a protein, known as interleukin-17A (IL-17A), found in immune cells, and in particular T cells that recognize commensals. When these cells sense an injury, they release IL-17A that is sensed by the damaged nerves and coordinate neuronal repair. Researchers from NIAID’s Laboratory of Host Immunity and Microbiome led the work in collaboration with NIH’s National Institute of Child Health and Human Development and the National Center for Complementary and Integrative Health. Researchers from Harvard Medical School also collaborated.
The findings add to the growing knowledge of how the microbiota – the trillions of beneficial microbes living harmoniously on our skin and inside our gut – bridge biological systems to benefit living beings. The group plans to continue its exploration of exactly how IL-17A communicates with the nervous system to repair damaged tissue and how these findings could lead to novel therapeutic approaches.
Reference:
M. Enamorado et al. Immunity to the microbiota promotes sensory neuron regeneration. Cell. DOI: https://doi.org/10.1016/j.cell.2022.12.037 (2023).