Tuberculosis Research Section
Established in 1995
Clifton Barry III, Ph.D.
Chief, Tuberculosis Research Section

Major Areas of Research
- Tuberculosis (TB) drug discovery
- Mechanism of action of anti-TB agents
- Drug resistance in Mycobacterium tuberculosis
- Chemical biology of the interaction of TB and humans
- Clinical trials of therapies in TB patients
Program Description
Tuberculosis (TB) is one of the leading infectious diseases in the world, with approximately one-third of the world’s population harboring the causative agent, Mycobacterium tuberculosis (Mtb). Though previously a disease associated with aristocratic societies, TB is now predominantly a third-world disease, particularly affecting Asian communities and sub-Saharan Africa. Mtb isolates are increasingly resistant to drug therapies: multidrug-resistant TB (MDR TB) or, more severely, extensively drug-resistant TB (XDR TB). As a consequence of these emerging strains, it is becoming increasingly apparent that novel drugs are necessary to combat Mtb infections.
Listen to Dr. Barry’s appearance on the TB Podcast, speaking about his involvement in the PREDICT TB study, the TB Drug Accelerator, and some of the most exciting drug developments in recent years.
Biography
Education
Ph.D., 1989, Cornell University
Dr. Clifton E. Barry III received his Ph.D. in organic and bioorganic chemistry in 1989 from Cornell University, studying the biosynthesis of complex natural products. Following postdoctoral research in the chemistry department at Johns Hopkins University (1989 to 1992), Dr. Barry joined the NIH Intramural Research Program at the NIAID Rocky Mountain Laboratories in Hamilton, Montana. In 1998, he was tenured as chief of the Tuberculosis Research Section (TBRS) in the Laboratory of Clinical Infectious Diseases.
TBRS is a multidisciplinary group of research scientists composed of biologists, chemists, and clinicians who share a common focus on tuberculosis (TB). TBRS projects focus on understanding the scientific issues that facilitate the development of drugs that will make a genuine difference in the outcome for TB patients globally. TBRS scientists are highly interactive worldwide in this endeavor. As a result of our outstanding collaborations, TBRS is the most highly cited TB research group in the world, according to Thomson Reuters. Dr Barry has authored over 300 publications in scientific literature. Working with scientists at PathoGenesis in Seattle, TBRS played a key role in the preclinical development program that led to PA-824 (Pretomanid, recently approved for treating drug-resistant TB). TBRS scientists conceived and conducted the Phase 2 clinical trial showing the utility of using linezolid for the treatment of patients suffering from extensively drug-resistant TB, now recommended by the World Health Organization. Working with scientists at Merck, TBRS has developed a safer oxazolidinone for TB that is currently in Phase 1 safety studies.
In addition to TBRS laboratories in Bethesda, TBRS works closely with colleagues at Stellenbosch University (SUN) and the University of Cape Town (UCT) in South Africa. Dr. Barry holds honorary appointments at SUN and UCT and has a laboratory in the Institute for Infectious Disease and Molecular Medicine at UCT.
Clinical Studies
Using Biomarkers to Predict TB Treatment Duration, NCT02821832
Selected Publications
Libardo MDJ, Duncombe CJ, Green SR, Wyatt PG, Thompson S, Ray PC, Ioerger TR, Oh S, Goodwin MB, Boshoff HIM, Barry CE 3rd. Resistance of Mycobacterium tuberculosis to indole 4-carboxamides occurs through alterations in drug metabolism and tryptophan biosynthesis. Cell Chem Biol. 2021 Aug 19;28(8):1180-1191.e20.
Xie YL, de Jager VR, Chen RY, Dodd LE, Paripati P, Via LE, Follmann D, Wang J, Lumbard K, Lahouar S, Malherbe ST, Andrews J, Yu X, Goldfeder LC, Cai Y, Arora K, Loxton AG, Vanker N, Duvenhage M, Winter J, Song T, Walzl G, Diacon AH, Barry CE 3rd. Fourteen-day PET/CT imaging to monitor drug combination activity in treated individuals with tuberculosis. Sci Transl Med. 2021 Feb 3;13(579):eabd7618.
Wang Q, Boshoff HIM, Harrison JR, Ray PC, Green SR, Wyatt PG, Barry CE 3rd. PE/PPE proteins mediate nutrient transport across the outer membrane of Mycobacterium tuberculosis. Science. 2020 Mar 6;367(6482):1147-1151.
Via LE, Weiner DM, Schimel D, Lin PL, Dayao E, Tankersley SL, Cai Y, Coleman MT, Tomko J, Paripati P, Orandle M, Kastenmayer RJ, Tartakovsky M, Rosenthal A, Portevin D, Eum SY, Lahouar S, Gagneux S, Young DB, Flynn JL, Barry CE 3rd.Differential virulence and disease progression following Mycobacterium tuberculosis complex infection of the common marmoset (Callithrix jacchus). Infect Immun. 2013 Aug;81(8):2909-19.
Lee M, Lee J, Carroll MW, Choi H, Min S, Song T, Via LE, Goldfeder LC, Kang E, Jin B, Park H, Kwak H, Kim H, Jeon HS, Jeong I, Joh JS, Chen RY, Olivier KN, Shaw PA, Follmann D, Song SD, Lee JK, Lee D, Kim CT, Dartois V, Park SK, Cho SN, Barry CE 3rd. Linezolid for treatment of chronic extensively drug-resistant tuberculosis. N Engl J Med. 2012 Oct 18;367(16):1508-18.
Via LE, Schimel D, Weiner DM, Dartois V, Dayao E, Cai Y, Yoon YS, Dreher MR, Kastenmayer RJ, Laymon CM, Carny JE, Flynn JL, Herscovitch P, Barry CE 3rd. Infection dynamics and response to chemotherapy in a rabbit model of tuberculosis using [18F]2-fluoro-deoxy-D-glucose positron emission tomography and computed tomography. Antimicrob Agents Chemother. 2012 Aug;56(8):4391-402.
Key LCID Collaborators
Key TBRS Collaborators
NCGC: NIH Chemical Genomics Center
OXU: Ben Davis’ laboratory at Oxford University
PITT: Joanne Flynn’s laboratory at the University of Pittsburgh
MRL: Merck Research Laboratories, Merck & Co., Inc. (2000 Galloping Hill Road, Kenilworth, New Jersey, USA 07033).
SUN: Gerhard Walzl’s laboratory at Stellenbosch University
GSK: GlaxoSmithKline, Tres Cantos, Spain
DDU: Drug discovery unit, University of Dundee
NCI: Barry O’Keefe’s Natural Products Branch of NCI
OCICB: NIAID’s Office of Cyber Infrastructure and Computational Biology
SBRI: David Sherman's laboratory at the Seattle Biomedical Research Institute
TASK: Andreas Diacon’s clinical team at TASK Applied Science in Stellenbosch
UMDNJA: David Alland’s laboratory at the University of Medicine and Dentistry of New Jersey
Tools/Resources
- BSL-3 High-throughput screening laboratory, Helena I. M. Boshoff, Staff Scientist
- TB Imaging Program (TBIP), Laura Via, Staff Scientist
Project Listing
TBRS works on myriad projects, ranging from drugs currently in clinical trials through cell and molecular biology and down to basic chemistry.
- Predict TB Clinical Trial
- PET/CT Imaging: NexGen EBA
- Chemical Genetics
- Mechanism of Action Studies
- TB Drug Accelerator
Predict TB Clinical Trial (SUN, TASK)
Shortening the duration of treatment for patients with drug sensitive tuberculosis from 6 to 4 months has been attempted many times in clinical trials but thus far all have failed. These failures reveal our incomplete understanding of factors driving the need for such extensive treatments. Consistently, trials have demonstrated that 80-85% of patients are successfully cured after 4 months of therapy, including the extensive set of studies from the British Medical Research Council (BMRC) in the 1970s and 1980, the Tuberculosis Research Unit (TBRU) treatment shortening study in non-cavitary patients who achieve early culture conversion, and the more recent treatment shortening trials using fluoroquinolones like REMoxTB. The current standard of care is to over-treat all patients for a total of 6-months to avoid relapse in a small subset of patients at higher risk for incompletely understood reasons.
For decades, clinical investigators have attempted to establish culture conversion as a predictor of treatment success. Despite the appealing logic, the real correlation of culture conversion as a surrogate endpoint has been consistently disappointing. In the REMoxTB trial, in particular, the intensive microbiological data collected revealed unambiguously that clearance of bacteria from the sputum did not sufficiently correlate with relapse risk to be a useful surrogate for durable cure. An important subset of patients, despite clearing their sputum of TB quickly and complying with all of their medications, still remained at high risk of relapsing with active disease after stopping treatment. Likewise, there are patients who clear their sputum of bacteria slowly that nonetheless go on to achieve durable cure. Intuitively this makes sense: only those bacteria at the surface of a cavity are directly open to the airways to seed the sputum. Yet this is not the full story as there are also heterogeneous lesions within each individual patient which respond differently to treatment with chemotherapy.
This protocol builds upon the historical trials and several successful small studies that suggest that directly monitoring lung pathology using (18F)- FDG PET/CT correlates better with treatment outcome than culture status. We will prospectively identify patients at low risk based on their baseline radiographic extent of disease, and further refine this risk score by evaluating the rate of resolution of the lung pathology (CT) and inflammation (PET) at one month as well as checking an end-of treatment GeneXpert test for the sustained presence of bacteria. Patients classified as low risk will be randomized to receive a shortened 4- month or a full 6-month course of therapy. If successful, this trial will both offer a badly needed alternative to culture status as a trial-level surrogate marker for outcome as well as provide critical information for preclinical and early clinical efforts to identify new agents and combinations with the potential to shorten therapy.
Hypothesis: A combination of radiographic characteristics at baseline, the rate of change of these features at one month, and markers of residual bacterial load at the end of treatment will identify patients with tuberculosis who are cured with 4 months (16 weeks) of standard treatment.
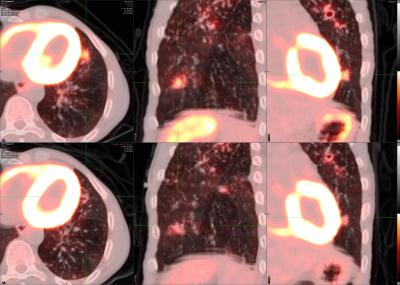
14 day PET/CT changes in a subject treated with quadruple drug standard of care treatment consisting of rifampicin, isoniazid, ethambutol and pyrazinamide. The top row depicts lesions immediately prior to beginning treatment and the bottom shows the aligned 14-day scan response.
Positron emission tomography (PET) scans illuminate areas of inflammation using radiolabeled tracers. Computed tomography (CT) takes high-resolution X-ray images of the body, allowing reconstruction of the 3-D structure of the scanned region. By simultaneously performing PET/CT scans, we can monitor Mtb infections in subjects over time to correlate lung structure and functional Mtb activity and measure the impact of new and existing chemotherapies.
PET/CT is an important endpoint in our on-going clinical trials, and we have used this as a surrogate endpoint for trials of new drugs for TB. We have developed quantitative segmentation and classification tools together with OCICB. We have developed the ability to monitor PET/CT changes in animal models of TB and are developing Mtb-specific PET probes to improve this technology.
Chemical Genetics—NCGC, NCI
(Left to right) Clif Barry, Helena Boshoff, Jessica Medrano (front), Jenna Andrews (back), Neha Malhotra, Alice Bell and Nelson Reyes collect samples from a bog in Maine in 2018.
One of the primary problems in new drug discovery for TB is the selection of appropriate targets that will have a therapeutic impact and kill the bacteria. This project seeks to screen for activity against whole cells of Mtb and then use these as starting points for target identification. By performing a variety of screens against Mtb under in vivo-relevant conditions (such as hypoxia, growth on lipid sources, and starvation), we have started assembling a “toolbox” of compounds that specifically inhibit growth of the pathogen under these conditions. One unique source of compounds is the “grey-layer” decomposition zone deep within sphagnum bogs (peat moss). This environment is highly acidic and microaerophilic, similar to that within human lungs where Mtb is thought to reside. Natural selection has endowed other microorganisms living in these nutrient-bare sphagnum areas with the ability to compete with endogenous slow-growing mycobacteria for these scarce nutrients by secreting secondary metabolites. Using microarray analysis and whole genome re-sequencing to identify the targets of these compounds, we can then identify potentially druggable leads.
TRS is also working with the Natural Products Branch of the NCI to screen and evaluate their new collection of over 1 million natural products contained in the NCI Program for Natural Product Discovery (NPNPD) Prefractionated Library.
TB Drug Accelerator
TRS works closely with the TB Drug Accelerator program of the Bill and Melinda Gates Foundation (https://globalhealthprogress.org/collaboration/tb-drug-accelerator-program/). We have a newly upgraded high-throughput screening facility within a Biosafety Level 3 containment suite that is routinely used to screen large compound libraries from collaborators (DDU, MRL). In addition, we conduct mechanism of action studies and formal hit assessment activities including a medicinal chemistry program to develop novel TB drugs.
We are also using a high throughput screening (HTS) robot to help quickly screen for potential drug targets that are revealed as necessary for Mtb survival under a range of growth conditions. These conditions, such as hypoxia, starvation, etc., mimic conditions that Mtb might encounter during the course of disease in a human host. We have adopted a quantitative HTS approach that screens all compounds as a titration series in order to get dose-response curves. The targets inhibited by these compound hits are subsequently identified by a variety of methods, including microarray analysis to compare profiles to our existing database of drug-induced transcriptional profiles, whole genome sequencing of resistant mutants, and macromolecular incorporation assays. Thus, the multitude of hits that are identified by HTS will enable the rapid discovery of multiple inhibitor-susceptible pathways that contain steps that are bottlenecks in the metabolism essential for survival under these conditions.
As part of this program, TRS recently concluded a partnership lead optimization program with MRL to develop a TB-specific oxazolidinone similar to linezolid without the known safety risks associated with mitochondrial toxicity. This program resulted in a candidate molecule that is about to begin clinical testing.
Research Group
TBRS is a multidisciplinary group composed of chemists, microbiologists, veterinarians, imaging scientists, and clinicians with a shared objective of improving chemotherapy for tuberculosis (TB) patients. Projects span chemical biology, medicinal chemistry, host-pathogen interactions, animal models of experimental chemotherapy, and experimental medicine clinical studies in TB patients.


